-
High Power and High Stakes: A Q&A on Machine Learning in Diagnostics
Heralding back to a landmark tuberculosis study in 2019, Analysis Group continues to deepen its application of machine learning to diagnostics.
Machine learning, broadly speaking, involves computer programs and algorithms that automatically improve their own performance at specific tasks through experience. Trained on existing data, they then learn to find highly complex new patterns on their own, without making assumptions about the relationships within the data.
The final step in a machine learning process is to apply the model to a real-world situation – such as screening patient data for indications of illness. In 2022, for example, an Analysis Group team contributed to a study demonstrating how machine learning could be used to screen the general population for post-traumatic stress disorder (PTSD). (See the callout below for examples of recent studies.)
In this panel, Managing Principals Annie Guérin and James Signorovitch and Principal Jimmy Royer discuss their experiences with using machine learning in diagnostic studies and their long-term visions for how this powerful tool could help reduce the global burden of various diseases.
Annie, machine learning is a hot topic, but your study of PTSD demonstrates an especially practical use of it. What’s unique about how your team used machine learning?
Ms. Guérin: When we know exactly what we’re looking for in a study, we don’t need machine learning. In our study, what we did know is that often veterans are screened for PTSD, but the general population is not.
That means many patients do not even know they could seek help for common symptoms, and physicians do not realize they have underserved patients. If it was very clear when patients have PTSD, underdiagnosis wouldn’t be as big a problem.
Machine learning is for just this sort of situation – where we don’t necessarily know everything to look for, so we ask, what else? Who are the patients most at risk of developing a certain event or a condition? Often, some factors are well known to be associated with higher risk of events, but are any of those factors more important than others for identifying patients?
In these kinds of situations, we may be missing other symptoms or factors that could be helpful for diagnosing a condition. If we don’t use machine learning, we wouldn’t know to include those variables in our model. Using a machine learning model, we let the data tell us what variables are important for building an algorithm to identify patients at risk of developing a certain condition, as we did with PTSD in this case.
So it’s not about pre-identifying which variables to include in the model, which would only reinforce any research biases. It’s letting the data tell us what to look for.
“Machine learning is for just this sort of situation – where we don’t necessarily know everything to look for, so we ask, what else? Who are the patients most at risk of developing a certain event or a condition?”– Annie Guérin
What do you think the larger impact of this study might be?
Ms. Guérin: The next step would be to put this tool in the hands of physicians. We’re not proposing a diagnosis. We’re saying, you should look at this set of patients more closely.
The stakes are quite high. For instance, we found that patients with likely undiagnosed PTSD had steeper rates of substance use, suicidal ideation, and other symptoms than patients who had been diagnosed with PTSD and so were more likely to be receiving treatment.
Knowing who is at risk can lead to an earlier diagnosis, which then improves the patient’s prognosis. Additionally, identifying the subpopulations more likely to benefit from diagnosis and treatment would also help payers with treatment authorization.
Jimmy, you did a study a few years ago that used machine learning to discern patients who might be likely to have multidrug-resistant or extensively drug-resistant tuberculosis. Why was this revolutionary at the time?
Dr. Royer: That study was groundbreaking for a couple of reasons. The data used in the model included information (in this case, about mutations) on segments of the genome of the bacteria sampled. An important aspect of that sample was the availability of the outcome of interest – namely, whether the bacteria were resistant, or susceptible, to a series of treatments. There is a significant volume of genomic data available to researchers, but being able to link a health outcome to the data, like we did here, is relatively rare.
The model itself was also novel. We used deep neural networks to analyze the data and predict resistance. The methodology was at the forefront of AI developments at the time.
How did you decide machine learning was the right tool for this study?
Dr. Royer: Genomic data is a peculiar type of data. Usually, in a dataset, you expect many more lines for patients (which typically are arrayed in rows in a dataset) than you have for variables (which are the columns). With genomic data you face the opposite issue, where so many variables can be extracted from the genome.
For this reason, we couldn’t use standard regression techniques. We investigated several machine learning models, tested out-of-sample performances, and ended up with deep neural networks. They’re much more flexible than a typical regression, which imposes restrictions on the form of the relationship between genomic data and its resistance profile. Most of these restrictions disappear when you use deep neural networks.
Like Annie said, the model itself will decide which variables or combinations are needed. In the genome or bacteria, a machine learning model will select which variables are likely to be important mutations in predicting treatment resistance.
“With AI and machine learning you can use atypical datasets, like the genomic composition of a bacteria or an image or picture, to predict a phenomenon. ”– Jimmy Royer
Pharmaceutical companies have used Analysis Group’s machine learning capabilities to generate real-world evidence and real-world data, working towards an end goal of helping patients. What other applications of machine learning are you excited about?
Dr. Royer: A sort of blue-sky use would be putting those capabilities in the hands of patients who do not have easy access to physicians. With AI and machine learning you can use atypical datasets, like the genomic composition of a bacteria or an image or picture, to predict a phenomenon.
Many people already have devices that can take a picture, such as a smartphone. Say someone has a skin condition. In an ideal world, you can imagine that person taking a picture of their skin using their smartphone and uploading it into a mobile application. The embedded machine-learning algorithm could analyze the picture and help the person decide whether they should see a doctor.
This is a very powerful way to leverage machine learning and predictive modeling in general. Such tools could help expand access to health care.
We are also working on machine learning models that can be integrated into an EMR [electronic medical record] for real-time decision support. The goal is to tailor them to different health care systems and settings, and make sure they’re both well calibrated and equitable to a diverse patient population.
James, you’ve spearheaded a number of machine learning studies. What kinds of benefits can machine learning deliver in these cases?
Dr. Signorovitch: For one, we’ve seen tremendous value from using machine learning to support diagnosis and prognosis for diseases that are rare and/or heterogenous, such as Sjögren’s syndrome, chronic urticaria, and muscular dystrophy.
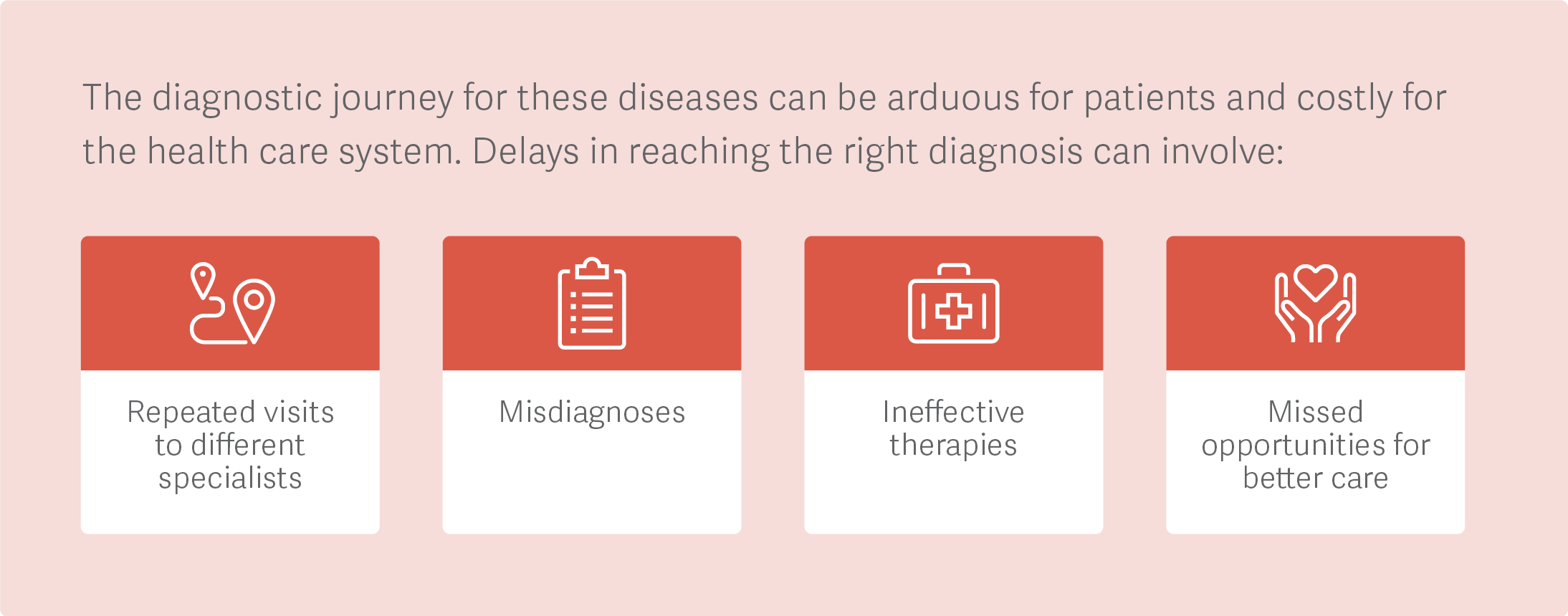
Unfortunately, when a disease can present different combinations of signs and symptoms, it is challenging for someone who’s not an expert to reach an accurate diagnosis – or even to know which expert to refer the patient to. And even an expert in a disease can struggle to make an accurate prognosis based only on their individual experience with patients.
Machine learning provides quantitative insights about diagnosis and prognosis that are based on many more patient encounters than any individual decision maker could directly experience. Ultimately, clinicians are still making the decisions, but they have more accurate, timely, and rigorous evidence to work with.
How does machine learning deal with the different kinds of data that characterize clinical studies?
Dr. Signorovitch: We’ve also seen value for machine learning in reducing the amount of information required to make a useful clinical prediction, essentially squeezing more information out of less data. This can be especially valuable when collecting data is burdensome or invasive for patients – for example, in tests of physical function that fatigue or stress the patient, or with invasive tissue assessments.
Beyond patient-level data, we have used machine learning models to draw insights from large bodies of text, such as scientific abstracts, or public regulatory documents from the FDA, the EMA [European Medicines Agency], and multiple HTA [health technology assessment] authorities.
When supporting the development of innovative new therapies, there is often a need to identify relevant precedents that might span diverse therapeutic areas. Machine learning and large language models, building on the elements that underlie tools such as ChatGPT, have been valuable in helping experts with deep experience more efficiently augment and pressure-test their thinking by drawing on vast and rapidly accumulating bodies of evidence.
What is next on the horizon?
Ms. Guérin: It will be important to develop very comprehensive data sources. Machine learning is a very powerful tool, but it’s only one method to address important research questions. Often, even if we have very robust, bespoke tools, we lack sufficient clinical data to which we can apply the tool.
Over time, more and more real-world data sources are becoming available. Such massive and complex datasets will necessitate machine learning models, and people who know how to use such models well.
“Machine learning provides quantitative insights about diagnosis and prognosis that are based on many more patient encounters than any individual decision maker could directly experience.”– James Signorovitch
Dr. Signorovitch: Validation is critical for machine learning applications in health care. An algorithm may perform well for a few data sources at a particular place and time, but this does not mean that it will always work everywhere. Validation and calibration need to be ongoing processes for real-world use.
We also need to recognize that real-world implementation – how machine learning tools are integrated with human expertise and workflows to impact decisions – can be equally or even more important than the quantitative performance of the algorithms themselves. We see increasing need for clinical utility and implementation science studies that evaluate the real-world impacts of integrating algorithms into health care decision-making.
The role of regulators, such as the FDA, in evaluating algorithms in health care is rapidly evolving, with recent draft guidance1 representing a major step forward. It is a sure bet, and a welcome development, that the FDA and many other decision makers will increasingly require more rigorous evidence of algorithm performance. ■
-
Applying Machine Learning in Clinical Studies
Analysis Group researchers have contributed to studies that have employed machine learning in a number of different disease areas. Some examples are below.
“Beyond multidrug resistance: Leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction,” M.L. Chen, A. Doddi, J. Royer, L. Freschi, M. Schito, M. Ezewudo, I.S. Kohane, A. Beam, and M. Farhat, EBioMedicine, Vol. 43 (2019)
“Early Predictors of Sjögren’s Syndrome: A Machine Learning Approach,” J. Signorovitch, I. Pivneva, W. Huber, and G. Capkun, Value in Health, Vol. 22, Supp. 2 (2019)
“Development of a Multivariable Proxy Model for Six-Minute Walk Distance (6MWD) in Duchenne Muscular Dystrophy (DMD) Using Machine Learning Methods,” N. Done, J. Iff, J. Signorovitch, D. Bertsimas, E. Henricson, and G. McDonald, Neurology, Vol. 94 (2020)
“Predicting Clinical Remission of Chronic Urticaria Using Random Survival Forests: Machine Learning Applied to Real-World Data,” I. Pivneva, M-M. Balp, Y. Geissbühler, T. Severin, S. Smeets, J. Signorovitch, J. Royer, et al., Dermatology and Therapy, Vol. 12 (2022)
“Development and evaluation of a predictive algorithm for unsatisfactory response among patients with pulmonary arterial hypertension using health insurance claims data,” M. Gauthier-Loiselle, Y. Tsang, P. Lefebvre, P. Agron, K.B. Lynum, L. Bennett, and S. Panjabi, Current Medical Research and Opinion, Vol. 38 (2022)
“A concisely recorded ambulatory assessment for enhancing real-world outcomes research in Duchenne muscular dystrophy: Development and validation,” A. Mayhew, J. Signorovitch, V. Straub, C. Marini Bettolo, R. Muni-Lofra, A. Manzur, V. Ayyar Gupta, V. Selby, and F. Muntoni, Neuromuscular Disorders, Vol. 32, Supp. 1, S66 (Oct. 2022)
“Validation of a composite prognostic score for time to loss of ambulation in Duchenne muscular dystrophy,” C. McDonald, H. Gordish-Dressman, J. Signorovitch, et al., Neuromuscular Disorders, Vol. 32, Supp. 1, S69 (Oct. 2022)
-
Endnote-
See, for example, the FDA’s Draft Guidance Document “Marketing Submission Recommendations for a Predetermined Change Control Plan for Artificial Intelligence/Machine Learning (AI/ML)-Enabled Device Software Functions” (April 2023).
-
This feature was published in May 2023.